The "Force of Mortality" -
reflections on the survival curve and the "ageing" process with
some implications for new directions in research.
Chadd Everone
{http://www.fis.org/public/obiterdicta/forceofmortality.html}
When Benjamin Gompertz published, in 1825,
his seminal work on the laws which seem to govern human mortality (1), he used the idea of the "force of mortality"
to convey the observation that something other than ordinary life-contingencies
was causing mortality after the age of 30. This observation became the
key-stone in the actuarial mathematics of the insurance industry as well
as in modern gerontology and life-extension science. The "force of
mortality" continues to be an apropos metaphor; and here, I
will draft some new perspectives on what might be the nature of that "force".
The Survival Curve of an Experimental
Animal Model
The C57BL/6j Mouse is an in-bred strain that
is probably the most widely used mouse in experimental biology. Because
it is long-lived and has a variety of chronic pathologies that are similar
to humans, it is believed to age and, therefore, is a good model for studies
in gerontology and life-extension. See: An Epistemology for Life-Extension
Science - The Experimental Animal Model - A Consensus Survey {http://www.fis.org/epistemology}.
The lineage of this mouse dates as follows. From Chinese through Japanese
and English "fanciers", who raised mice as an aesthetic hobby,
this and almost all inbred strains of mice used in experimental science
today were developed between 1903-1915 at Abbie Lathrop's mouse farm in
Granby, Maryland. One was labeled "C57". Through Clarence Little,
a "Black" sub-line was developed between 1921 and 1937. And from
that, another sub-line, called "6", was developed at Jackson Laboratory in 1947 - thus, becoming
named as C57BL/6j.
Since 1947, this strain has been bred (sister
and brother) for countless generations by the central breeding laboratory
(Jackson) and by other breeding colonies and investigators all over the
world.
The strain is extremely stable and, given
reasonably adequate husbandry, the survival curve has been virtually identical
in hundreds of life-span studies over some 6 decades by hundreds of investigators
in many different locations and under varying conditions of nutrition,
sanitation, and other environmental conditions. Below, the first survival
curve is from the breeding laboratory, Jackson Laboratory, done in 1979.
The second survival curve is from the colony of this author from an experiment
almost 10 years later in 1987 and done under quite different conditions.
As can be seen, the survival curves and mortality distributions are virtually
identical.
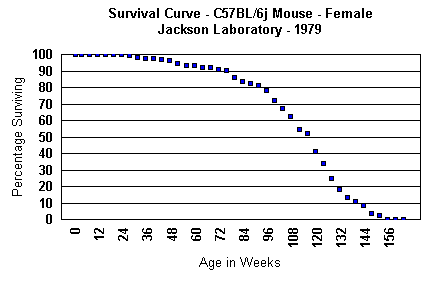
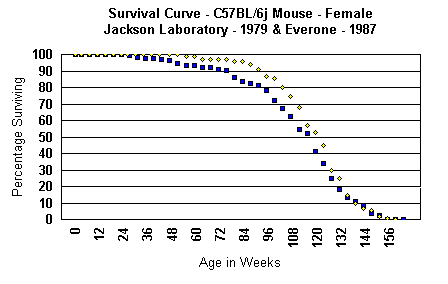
Having been inbred for so many generations,
the C57BL/6 stain is so homogeneous that all individuals are, genomically,
virtually identical - they are essentially clones. An experimental group
of 100 or 1,000 animals is like having 100 or 1,000 identical individuals.
Thus, one might expect that all individuals would have the same diseases
and closely identical lengths of survival; but, in fact, they do not. From
the graphs, above, it can be seen that 25% live out to 125 - 164 weeks,
while 50% live 116 weeks, and 25% do not live more than 97 weeks. These
percentages are practically invariable between different, experimental
colonies.
Several observations pertain. Given that
these mice are genetically identical, it is remarkable that there is such
a spread of individual survivability. Also, it is remarkable that this
spread in survivability is so uniform and consistent over decades of time
and under so many environmental conditions. It is as if the variation in
survivability and the distribution of its array is genetically programmed
- but not at the level of the genome itself.
When I discussed this phenomenon with a geneticist
at UC Berkeley, who is now conducting a large-scale gene knock-out study
on this strain of mouse, without any hesitation, he said: "Its obvious,
it is all determined during early development!" In March 2002, Craig Venter, the founder of Celera
and one of the key scientist in the Human Genome Project, delivered the
Hitchcock Lectures at UCB; and before one of the presentations, I presented
this for his consideration. He said: "That just shows that it is not
all genomic." But when it was pointed out that the survival differential
was so constant, he thought for a moment and then said: "To tell you
the truth, I do not have the slightest idea." (His candor and objectivity
was appreciated and commendable.) Again, in March 2002, Wolf
Reik gave a presentation at Lawrence Berkeley Laboratory about
his work on "Dynamic Reprogramming of DNA Methylation in the Early
Mouse Embryo", and this issue was brought to his attention. He had
never considered it before but suggested that an experiment might be designed
to analyze DNA methylation of a cohort to determine the differences in
the reprogramming of different individual genomes and then carry-out the
experiment to the full-term life-span and see if there might be any correlation.
The Survival Curve of Human Populations
Unlike
the experimental animal model above, human populations are very heterogeneous;
and our environmental variables are radically different and disparate.
Regardless, our survival curve and distribution of mortality are similar.
The exponential rate of mortality after age 30 is held to be caused by
"ageing" - i.e., Gompertz' "force of mortality".
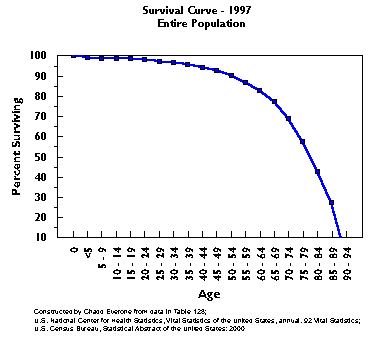
Curiously, if you take all of the people
who die from one particular chronic disease and create a survival curve
for only that group, you obtain similar survival curves.